Get a set of feature data for a spatial grid
get_features.RdThis is a wrapper of get_bathymetry(),
get_seamounts_buffered(), get_knolls(), get_geomorphology(),
get_coral_habitat(), and get_enviro_zones(). See the individual
functions for details.
Usage
get_features(
spatial_grid = NULL,
raw = FALSE,
features = c("bathymetry", "seamounts", "knolls", "geomorphology", "corals",
"enviro_zones"),
seamount_buffer = 30000,
antipatharia_threshold = 22,
octocoral_threshold = 2,
enviro_clusters = NULL,
max_enviro_clusters = 6,
antimeridian = NULL
)Arguments
- spatial_grid
sforterra::rast()grid, e.g. created usingget_grid(). Alternatively, if raw data is required, ansfpolygon can be provided, e.g. created usingget_boundary(), and setraw = TRUE.- raw
logicalif TRUE,spatial_gridshould be ansfpolygon, and the raw feature data in that polygon(s) will be returned. Note that this will be a list object, since raster andsfdata may be returned.- features
a vector of feature names, can include: "bathymetry", "seamounts", "knolls", "geomorphology", "corals", "enviro_zones"
- seamount_buffer
numeric; the distance from the seamount peak to include in the output. Distance should be in the same units as the area_polygon or spatial_grid provided, use e.g.sf::st_crs(spatial_grid, parameters = TRUE)$units_gdalto check what units your planning grid or area polygon is in (works for raster as well as sf objects)- antipatharia_threshold
numericbetween 0 and 100; the threshold value for habitat suitability for antipatharia corals to be considered present (default is 22, as defined in Yesson et al., 2017)- octocoral_threshold
numericbetween 0 and 7; the threshold value for how many species (of 7) should be predicted present in an area for octocorals to be considered present (default is 2)- enviro_clusters
numeric; the number of environmental zones to cluster the data into - to be used when a clustering algorithm is not necessary (default is NULL)- max_enviro_clusters
numeric; the maximum number of environmental zones to try when using the clustering algorithm (default is 8)- antimeridian
Does
spatial_gridspan the antimeridian? If so, this should be set toTRUE, otherwise set toFALSE. If set toNULL(default) the function will try to check ifspatial_gridspans the antimeridian and set this appropriately.
Value
If raw = TRUE, a list of feature data is returned (mixed raster and
sf objects). If a spatial_grid is supplied, a multi-layer raster or
sf object of gridded data is returned, depending on the spatial_grid
format.
Examples
# Grab EEZ data first
bermuda_eez <- get_boundary(name = "Bermuda")
# Get raw data for Bermuda's EEZ
raw_data <- get_features(spatial_grid = bermuda_eez, raw = TRUE)
#> Getting depth zones...
#> Bathymetry data already downloaded, using cached version
#> Getting seamount data...
#> Spherical geometry (s2) switched off
#> although coordinates are longitude/latitude, st_intersection assumes that they
#> are planar
#> Warning: attribute variables are assumed to be spatially constant throughout all geometries
#> Warning: st_buffer does not correctly buffer longitude/latitude data
#> dist is assumed to be in decimal degrees (arc_degrees).
#> Spherical geometry (s2) switched on
#> Getting knoll data...
#> Spherical geometry (s2) switched off
#> although coordinates are longitude/latitude, st_intersection assumes that they
#> are planar
#> Warning: attribute variables are assumed to be spatially constant throughout all geometries
#> Spherical geometry (s2) switched on
#> Getting geomorphology data...
#> Getting coral data...
#> Getting environmental zones data... This could take several minutes
# Get feature data in a spatial grid
bermuda_grid <- get_grid(boundary = bermuda_eez, crs = '+proj=laea +lon_0=-64.8108333 +lat_0=32.3571917 +datum=WGS84 +units=m +no_defs', resolution = 20000)
#set seed for reproducibility in the get_enviro_zones() function
set.seed(500)
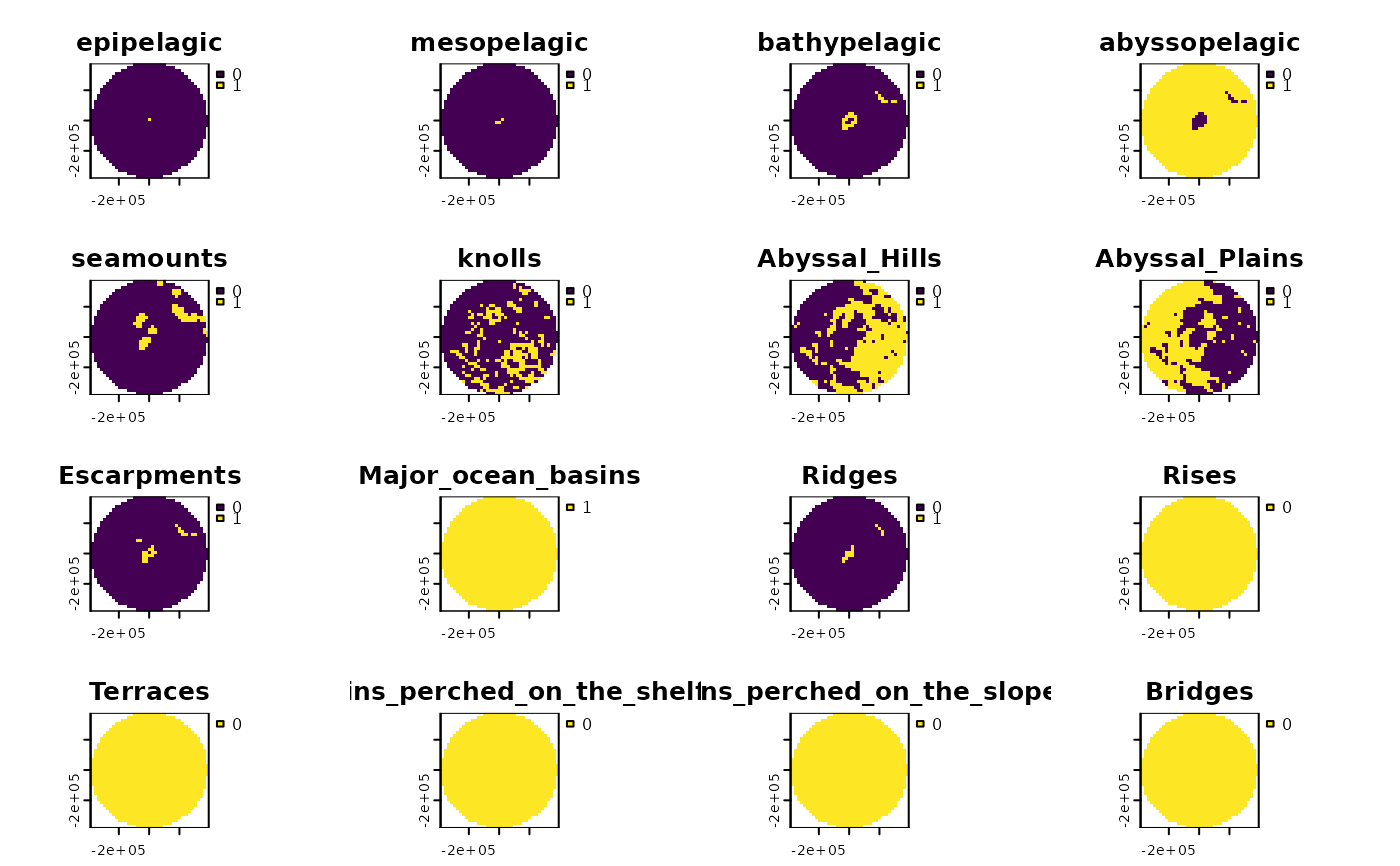
features_gridded <- get_features(spatial_grid = bermuda_grid)
#> Getting depth zones...
#> Bathymetry data already downloaded, using cached version
#> Getting seamount data...
#> Spherical geometry (s2) switched off
#> although coordinates are longitude/latitude, st_intersection assumes that they
#> are planar
#> Warning: attribute variables are assumed to be spatially constant throughout all geometries
#> Spherical geometry (s2) switched on
#> Getting knoll data...
#> Spherical geometry (s2) switched off
#> although coordinates are longitude/latitude, st_intersection assumes that they
#> are planar
#> Warning: attribute variables are assumed to be spatially constant throughout all geometries
#> Spherical geometry (s2) switched on
#> Getting geomorphology data...
#> Getting coral data...
#> Getting environmental zones data... This could take several minutes
terra::plot(features_gridded)