Create environmental regions for area of interest
get_enviro_regions.RdThis function gets Bio-Oracle
environmental data for the spatial grid and can then create environmental
regions using k-means clustering. The idea for the clustering comes from
Magris et al. 2020. The number of
environmental regions can be specified directly, using num_clusters, but
the function can also find the 'optimal' number of clusters using the
NbClust() from the NbClust package.
Usage
get_enviro_regions(
spatial_grid = NULL,
raw = FALSE,
enviro_regions = TRUE,
show_plots = FALSE,
num_clusters = NULL,
max_num_clusters = 6,
antimeridian = NULL,
sample_size = 5000,
num_samples = 5,
num_cores = 1,
custom_seed = 1234
)Arguments
- spatial_grid
sforterra::rast()grid, e.g. created usingget_grid(). Alternatively, if raw data is required, ansfpolygon can be provided, e.g. created usingget_boundary(), and setraw = TRUE.- raw
logicalif TRUE,spatial_gridshould be ansfpolygon, and the raw Bio-Oracle environmental data in that polygon(s) will be returned, unlessenviro_regions = TRUE, in which case the raw data will be classified into environmental regions- enviro_regions
logicalif TRUE, environmental regions will be created. If FALSE the gridded Bio-Oracle data will be returned- show_plots
logical; whether to show boxplots for each environmental variable in each environmental region (default is FALSE)- num_clusters
numeric; the number of environmental regions to cluster the data into - to be used when a clustering algorithm is not necessary (default is NULL)- max_num_clusters
numeric; the maximum number of environmental regions to try when using the clustering algorithm (default is 6)- antimeridian
Does
spatial_gridspan the antimeridian? If so, this should be set toTRUE, otherwise set toFALSE. If set toNULL(default) the function will try to check ifspatial_gridspans the antimeridian and set this appropriately.- sample_size
numeric; default is 5000. Larger sample sizes will quickly consume memory (>10GB) so should be used with caution.- num_samples
numeric; default is 5, which resulted in good consensus on the optimal number of clusters in testing.- num_cores
numeric; default 1. Multi-core sampling is supported if the packageparallelis installed, but be aware than increasing the number of cores will also increase the memory required.- custom_seed
numeric; default1234, but a custom seed can be supplied if desired.
Value
If enviro_regions = FALSE, Bio-Oracle data in the spatial_grid
supplied, or in the original raster file resolution if raw = TRUE. If
enviro_regions = TRUE a multi-layer raster or an sf object with one
environmental region in each column/ layer is returned, depending on the
spatial_grid format. If enviro_regions = TRUE and raw = TRUE (in
which case spatial_grid should be an sf polygon), the raw Bio-Oracle
data is classified into environmental zones.
Details
The environmental data used in the clustering are all sea surface measurements over the period 2010 - 2020:
Chlorophyll concentration (mean, mg/ m3)
Dissolved oxygen concentration (mean)
Nitrate concentration (mean, mmol/ m3)
pH (mean)
Phosphate concentration (mean, mmol/ m3)
total Phytoplankton (primary productivity; mean, mmol/ m3)
Salinity (mean)
Sea surface temperature (max, degree C)
Sea surface temperature (mean, degree C)
Sea surface temperature (min, degree C)
Silicate concentration (mean, mmol/ m3)
For full details of the Bio-Oracle data see Assis et al. 2024.
When the number of planning units/ cells for clustering exceeds ~ 10,000,
the amount of computer memory required to find the optimal number of
clusters using NbClust::NbClust() exceeds 10GB, so repeated sampling is
used to find a consensus number of clusters. Sensible defaults for
NbClust() are provided, namely sample_size = 5000, num_samples = 5,
max_num_clusters = 6 but can be customised if desired, though see the
parameter descriptions below for some words of warning. Parallel processing
is offered by specifying num_cores >1 (must be an integer), though the
package parallel must be installed (it is included in most R
installations). To find the number of available cores on your systems run
parallel::detectCores().
Examples
# Get EEZ data first
bermuda_eez <- get_boundary(name = "Bermuda")
#> Cache is fresh. Reading: /tmp/Rtmpr9MFzz/eez-2205f12f/eez.shp
#> (Last Modified: 2025-01-14 22:28:09.583216)
# Get raw Bio-Oracle environmental data for Bermuda
enviro_data <- get_enviro_regions(spatial_grid = bermuda_eez, raw = TRUE, enviro_regions = FALSE)
#> Selected dataset chl_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/chl_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: chl_mean
#> Selected dataset o2_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/o2_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: o2_mean
#> Selected dataset no3_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/no3_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: no3_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_min
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_max
#> Selected dataset ph_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/ph_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: ph_mean
#> Selected dataset po4_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/po4_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: po4_mean
#> Selected dataset so_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/so_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: so_mean
#> Selected dataset si_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/si_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: si_mean
#> Selected dataset phyc_baseline_2000_2020_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/phyc_baseline_2000_2020_depthsurf.html
#> Selected 1 variables: phyc_mean
terra::plot(enviro_data)
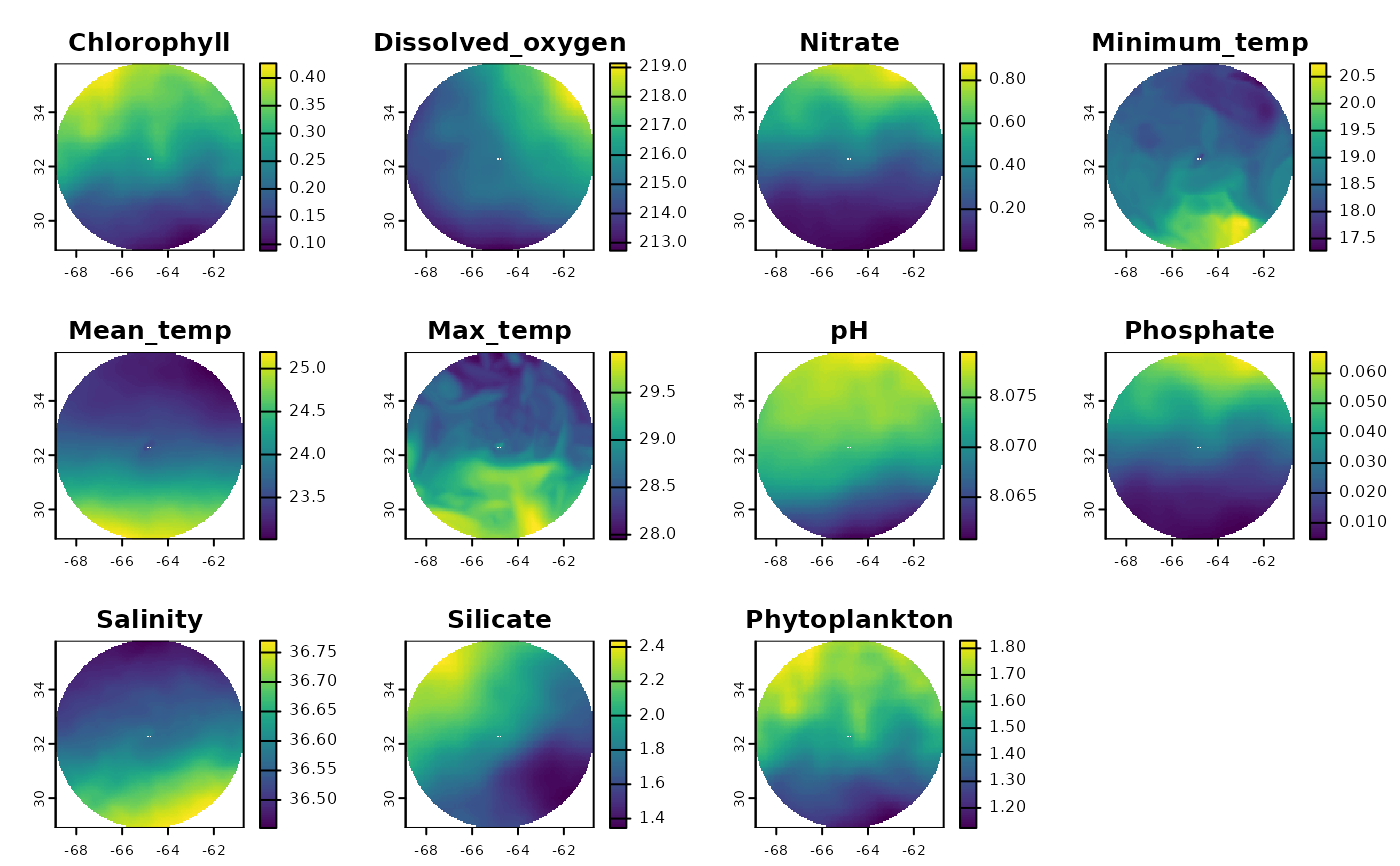 # Get gridded Bio-Oracle data for Bermuda:
bermuda_grid <- get_grid(boundary = bermuda_eez, crs = '+proj=laea +lon_0=-64.8108333 +lat_0=32.3571917 +datum=WGS84 +units=m +no_defs', resolution = 20000)
enviro_data_gridded <- get_enviro_regions(spatial_grid = bermuda_grid, raw = FALSE, enviro_regions = FALSE)
#> Selected dataset chl_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/chl_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: chl_mean
#> Selected dataset o2_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/o2_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: o2_mean
#> Selected dataset no3_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/no3_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: no3_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_min
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_max
#> Selected dataset ph_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/ph_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: ph_mean
#> Selected dataset po4_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/po4_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: po4_mean
#> Selected dataset so_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/so_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: so_mean
#> Selected dataset si_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/si_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: si_mean
#> Selected dataset phyc_baseline_2000_2020_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/phyc_baseline_2000_2020_depthsurf.html
#> Selected 1 variables: phyc_mean
terra::plot(enviro_data_gridded)
# Get gridded Bio-Oracle data for Bermuda:
bermuda_grid <- get_grid(boundary = bermuda_eez, crs = '+proj=laea +lon_0=-64.8108333 +lat_0=32.3571917 +datum=WGS84 +units=m +no_defs', resolution = 20000)
enviro_data_gridded <- get_enviro_regions(spatial_grid = bermuda_grid, raw = FALSE, enviro_regions = FALSE)
#> Selected dataset chl_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/chl_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: chl_mean
#> Selected dataset o2_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/o2_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: o2_mean
#> Selected dataset no3_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/no3_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: no3_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_min
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_max
#> Selected dataset ph_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/ph_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: ph_mean
#> Selected dataset po4_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/po4_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: po4_mean
#> Selected dataset so_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/so_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: so_mean
#> Selected dataset si_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/si_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: si_mean
#> Selected dataset phyc_baseline_2000_2020_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/phyc_baseline_2000_2020_depthsurf.html
#> Selected 1 variables: phyc_mean
terra::plot(enviro_data_gridded)
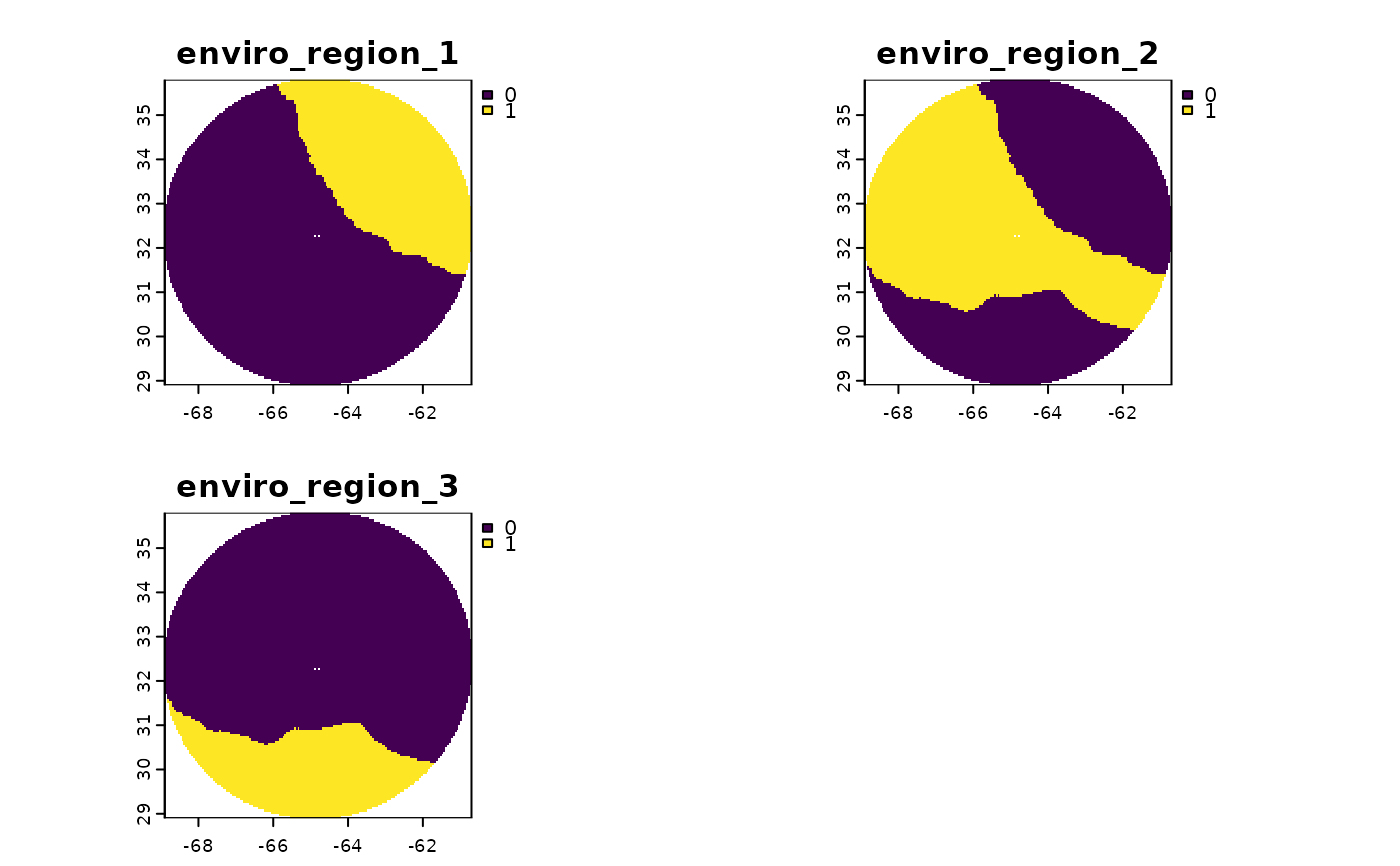
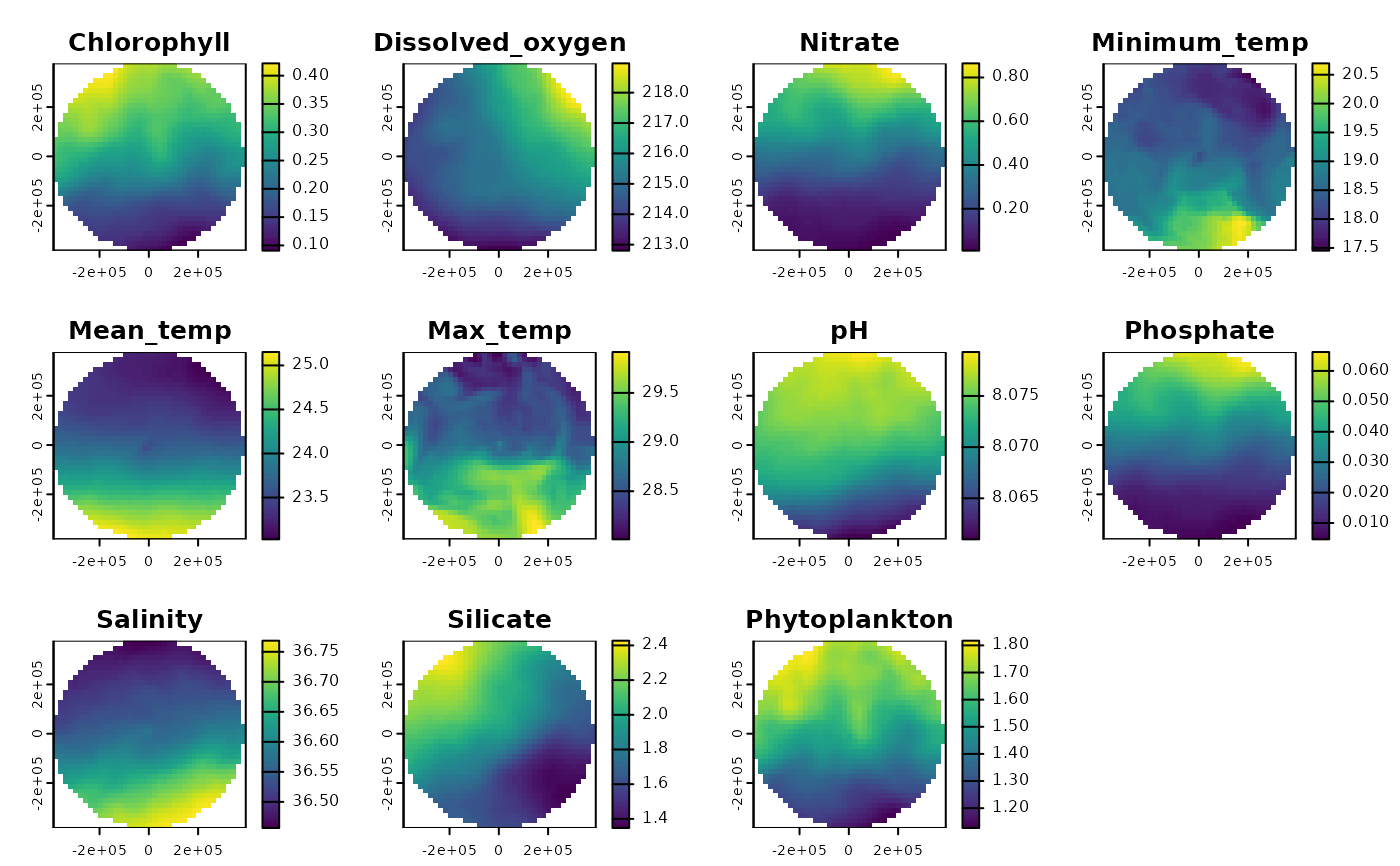 # Get 3 environmental regions for Bermuda
bermuda_enviro_regions <- get_enviro_regions(spatial_grid = bermuda_grid, raw = FALSE, enviro_regions = TRUE, num_clusters = 3)
#> Selected dataset chl_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/chl_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: chl_mean
#> Selected dataset o2_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/o2_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: o2_mean
#> Selected dataset no3_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/no3_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: no3_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_min
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_max
#> Selected dataset ph_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/ph_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: ph_mean
#> Selected dataset po4_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/po4_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: po4_mean
#> Selected dataset so_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/so_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: so_mean
#> Selected dataset si_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/si_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: si_mean
#> Selected dataset phyc_baseline_2000_2020_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/phyc_baseline_2000_2020_depthsurf.html
#> Selected 1 variables: phyc_mean
terra::plot(bermuda_enviro_regions)
# Get 3 environmental regions for Bermuda
bermuda_enviro_regions <- get_enviro_regions(spatial_grid = bermuda_grid, raw = FALSE, enviro_regions = TRUE, num_clusters = 3)
#> Selected dataset chl_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/chl_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: chl_mean
#> Selected dataset o2_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/o2_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: o2_mean
#> Selected dataset no3_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/no3_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: no3_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_min
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_max
#> Selected dataset ph_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/ph_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: ph_mean
#> Selected dataset po4_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/po4_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: po4_mean
#> Selected dataset so_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/so_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: so_mean
#> Selected dataset si_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/si_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: si_mean
#> Selected dataset phyc_baseline_2000_2020_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/phyc_baseline_2000_2020_depthsurf.html
#> Selected 1 variables: phyc_mean
terra::plot(bermuda_enviro_regions)
 # Can also create environmental regions from the raw Bio-Oracle data using setting raw = TRUE and enviro_regions = TRUE. In this case, the `spatial_grid` should be a polygon of the area you want the data for
bermuda_enviro_regions2 <- get_enviro_regions(spatial_grid = bermuda_eez, raw = TRUE, enviro_regions = TRUE, num_clusters = 3)
#> Selected dataset chl_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/chl_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: chl_mean
#> Selected dataset o2_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/o2_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: o2_mean
#> Selected dataset no3_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/no3_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: no3_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_min
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_max
#> Selected dataset ph_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/ph_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: ph_mean
#> Selected dataset po4_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/po4_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: po4_mean
#> Selected dataset so_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/so_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: so_mean
#> Selected dataset si_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/si_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: si_mean
#> Selected dataset phyc_baseline_2000_2020_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/phyc_baseline_2000_2020_depthsurf.html
#> Selected 1 variables: phyc_mean
terra::plot(bermuda_enviro_regions2)
# Can also create environmental regions from the raw Bio-Oracle data using setting raw = TRUE and enviro_regions = TRUE. In this case, the `spatial_grid` should be a polygon of the area you want the data for
bermuda_enviro_regions2 <- get_enviro_regions(spatial_grid = bermuda_eez, raw = TRUE, enviro_regions = TRUE, num_clusters = 3)
#> Selected dataset chl_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/chl_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: chl_mean
#> Selected dataset o2_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/o2_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: o2_mean
#> Selected dataset no3_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/no3_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: no3_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_min
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_mean
#> Selected dataset thetao_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/thetao_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: thetao_max
#> Selected dataset ph_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/ph_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: ph_mean
#> Selected dataset po4_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/po4_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: po4_mean
#> Selected dataset so_baseline_2000_2019_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/so_baseline_2000_2019_depthsurf.html
#> Selected 1 variables: so_mean
#> Selected dataset si_baseline_2000_2018_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/si_baseline_2000_2018_depthsurf.html
#> Selected 1 variables: si_mean
#> Selected dataset phyc_baseline_2000_2020_depthsurf.
#> Dataset info available at: http://erddap.bio-oracle.org/erddap/griddap/phyc_baseline_2000_2020_depthsurf.html
#> Selected 1 variables: phyc_mean
terra::plot(bermuda_enviro_regions2)