Manipulate Raster Data
Last updated on 2024-08-19 | Edit this page
Overview
Questions
- How can I crop raster objects to vector objects, and extract the summary of raster pixels?
Objectives
- Crop a raster to the extent of a vector layer.
- Extract values from a raster that correspond to a vector file overlay.
This episode explains how to crop a raster using the extent of a vector layer. We will also cover how to extract values from a raster that occur within a set of polygons, or in a buffer (surrounding) region around a set of points.
Let’s start by loading the libraries and data we need for this episode:
R
library(sf)
library(terra)
library(ggplot2)
library(dplyr)
R
point_HARV <-
st_read("data/NEON-DS-Site-Layout-Files/HARV/HARVtower_UTM18N.shp")
# If you are getting an error, check your file path:
# You might need change your file path to:
# "data/2009586/NEON-DS-Site-Layout-Files/HARV/HARVtower_UTM18N.shp"
aoi_boundary_HARV <-
st_read("data/NEON-DS-Site-Layout-Files/HARV/HarClip_UTMZ18.shp")
# If you are getting an error, check your file path:
# You might need change your file path to:
# "data/2009586/NEON-DS-Site-Layout-Files/HARV/HarClip_UTMZ18.shp"
# CHM data, and convert it to a dataframe
CHM_HARV <-
rast("data/NEON-DS-Airborne-Remote-Sensing/HARV/CHM/HARV_chmCrop.tif")
# If you are getting an error, check your file path:
# You might need change your file path to:
# "data/2009586/NEON-DS-Airborne-Remote-Sensing/HARV/CHM/HARV_chmCrop.tif"
CHM_HARV_df <- as.data.frame(CHM_HARV, xy = TRUE)
R
plot_locations_sp_HARV <- st_read("data/cleaned-data/PlotLocations_HARV.shp")
Crop a Raster to Vector Extent
We often work with spatial layers that have different spatial extents. The spatial extent of a vector layer or R spatial object represents the geographic “edge” or location that is the furthest north, south east and west. Thus it represents the overall geographic coverage of the spatial object.
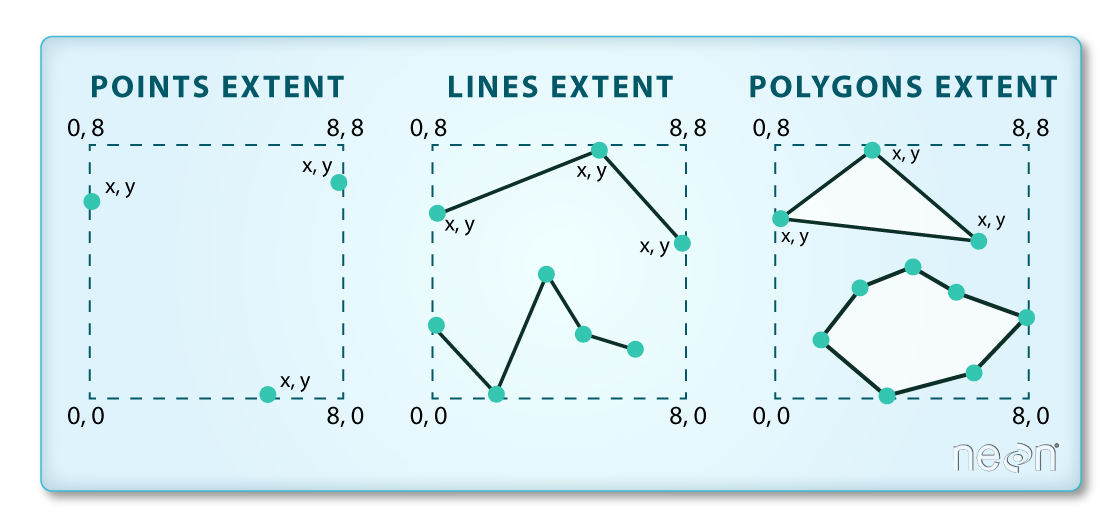 Image Source: National Ecological
Observatory Network (NEON)
Image Source: National Ecological
Observatory Network (NEON)
The graphic below illustrates the extent of several of the spatial layers that we have worked with in this workshop:
- Area of interest (AOI) – blue
- Roads and trails – purple
- Vegetation plot locations (marked with white dots)– black
- A canopy height model (CHM) in GeoTIFF format – green
Frequent use cases of cropping a raster file include reducing file size and creating maps. Sometimes we have a raster file that is much larger than our study area or area of interest. It is often more efficient to crop the raster to the extent of our study area to reduce file sizes as we process our data. Cropping a raster can also be useful when creating pretty maps so that the raster layer matches the extent of the desired vector layers.
Crop a Raster Using Vector Extent
We can use the crop() function to crop a raster to the
extent of another spatial object. To do this, we need to specify the
raster to be cropped and the spatial object that will be used to crop
the raster. R will use the extent of the spatial object as
the cropping boundary.
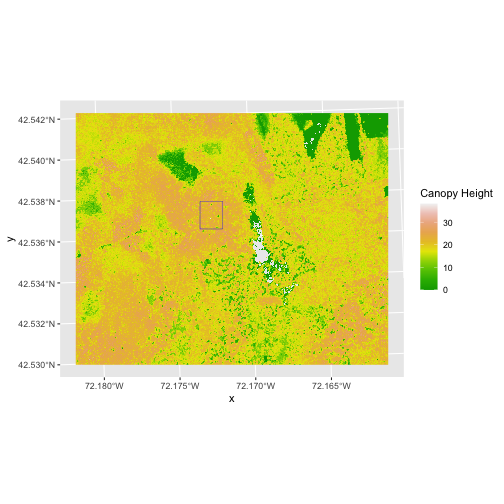
To illustrate this, we will crop the Canopy Height Model (CHM) to
only include the area of interest (AOI). Let’s start by plotting the
full extent of the CHM data and overlay where the AOI falls within it.
The boundaries of the AOI will be colored blue, and we use
fill = NA to make the area transparent.
R
ggplot() +
geom_raster(data = CHM_HARV_df,
mapping = aes(x = x, y = y, fill = HARV_chmCrop)) +
scale_fill_gradientn(colors = terrain.colors(10)) +
geom_sf(data = aoi_boundary_HARV, color = "blue", fill = NA) +
labs(fill = "Canopy Height") +
coord_sf()
Now that we have visualized the area of the CHM we want to subset, we
can perform the cropping operation. We are going to crop()
function from the raster package to create a new object with only the
portion of the CHM data that falls within the boundaries of the AOI.
R
CHM_HARV_Cropped <- crop(x = CHM_HARV, y = aoi_boundary_HARV)
Now we can plot the cropped CHM data, along with a boundary box
showing the full CHM extent. However, remember, since this is raster
data, we need to convert to a data frame in order to plot using
ggplot. To get the boundary box from CHM, the
st_bbox() will extract the 4 corners of the rectangle that
encompass all the features contained in this object. The
st_as_sfc() converts these 4 coordinates into a polygon
that we can plot:
R
CHM_HARV_Cropped_df <- as.data.frame(CHM_HARV_Cropped, xy = TRUE)
boundary <- st_as_sfc(st_bbox(CHM_HARV))
ggplot() +
geom_sf(data = boundary, fill = "green",
color = "green", alpha = .2) +
geom_raster(data = CHM_HARV_Cropped_df,
mapping = aes(x = x, y = y, fill = HARV_chmCrop)) +
scale_fill_gradientn(colors = terrain.colors(10)) +
labs(fill = "Canopy Height") +
coord_sf()
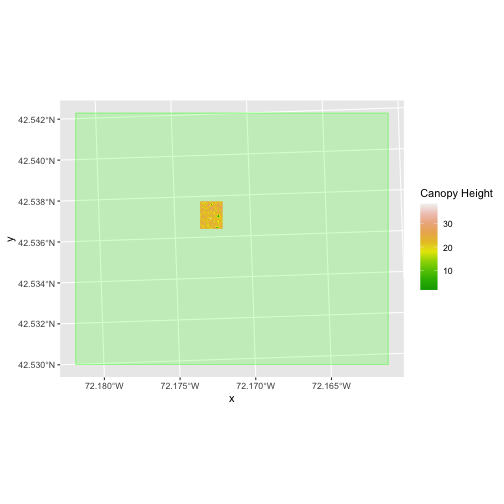
The plot above shows that the full CHM extent (plotted in green) is
much larger than the resulting cropped raster. Our new cropped CHM now
has the same extent as the aoi_boundary_HARV object that
was used as a crop extent (blue border below).
R
ggplot() +
geom_raster(data = CHM_HARV_Cropped_df,
mapping = aes(x = x, y = y, fill = HARV_chmCrop)) +
geom_sf(data = aoi_boundary_HARV, color = "blue", fill = NA) +
scale_fill_gradientn(colors = terrain.colors(10)) +
labs(fill = "Canopy Height") +
coord_sf()
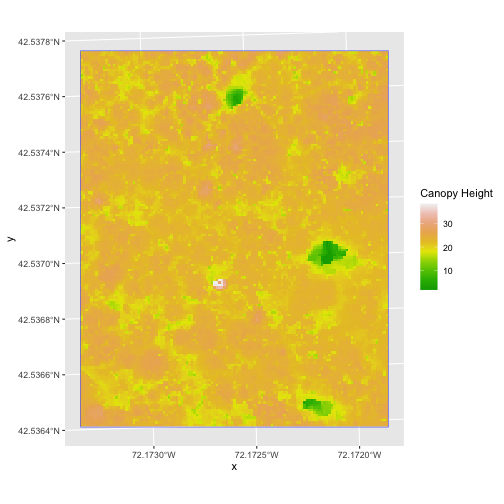
R
CHM_plots_HARVcrop <- crop(x = CHM_HARV, y = plot_locations_sp_HARV)
CHM_plots_HARVcrop_df <- as.data.frame(CHM_plots_HARVcrop, xy = TRUE)
ggplot() +
geom_raster(data = CHM_plots_HARVcrop_df,
mapping = aes(x = x, y = y, fill = HARV_chmCrop)) +
scale_fill_gradientn(colors = terrain.colors(10)) +
geom_sf(data = plot_locations_sp_HARV) +
labs(fill = "Canopy Height") +
coord_sf()
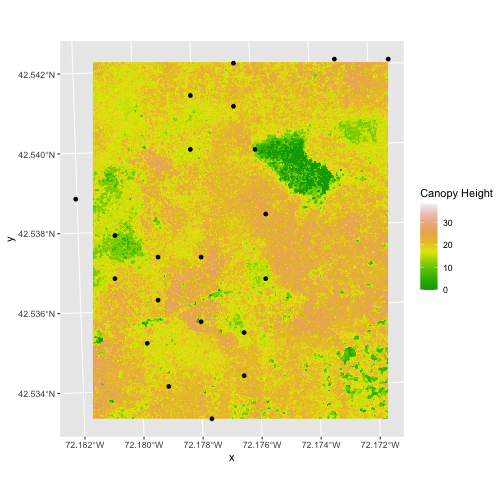
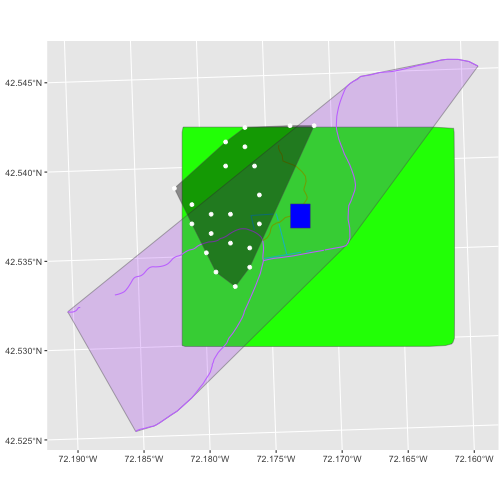
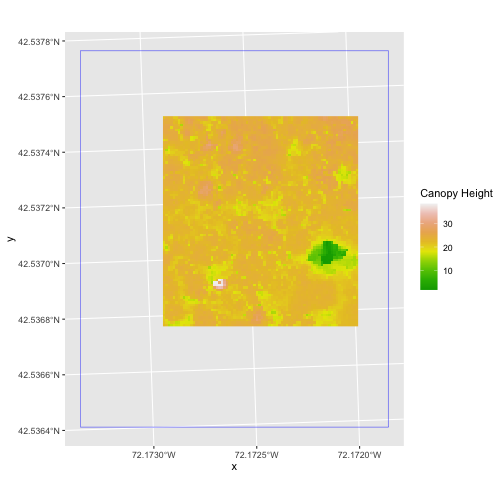
In the plot above, created in the challenge, all the vegetation plot locations (black dots) appear on the Canopy Height Model raster layer except for one. One is situated on the blank space to the left of the map. Why?
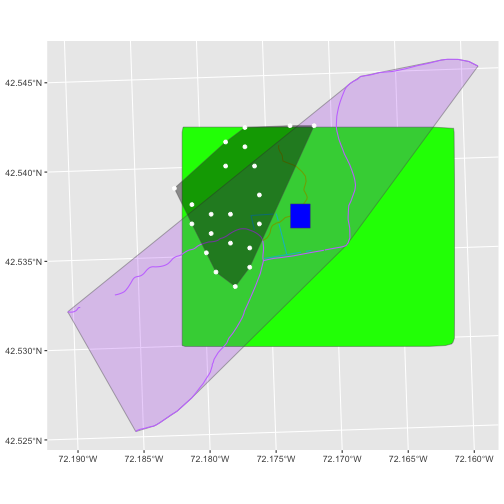
A modification of the first figure in this episode is below, showing
the relative extents of all the spatial objects. Notice that the extent
for our vegetation plot layer (black) extends further west than the
extent of our CHM raster (bright green). The crop()
function will make a raster extent smaller, it will not expand the
extent in areas where there are no data. Thus, the extent of our
vegetation plot layer will still extend further west than the extent of
our (cropped) raster data (dark green).
Define an Extent
So far, we have used a vector layer to crop the extent of a raster
dataset. Alternatively, we can also the ext() function to
define an extent to be used as a cropping boundary. This creates a new
object of class extent. Here we will provide the ext()
function our xmin, xmax, ymin, and ymax (in that order).
R
new_extent <- ext(732161.2, 732238.7, 4713249, 4713333)
class(new_extent)
OUTPUT
[1] "SpatExtent"
attr(,"package")
[1] "terra"Once we have defined our new extent, we can use the
crop() function to crop our raster to this extent
object.
R
CHM_HARV_manual_cropped <- crop(x = CHM_HARV, y = new_extent)
To plot this data using ggplot() we need to convert it
to a dataframe.
R
CHM_HARV_manual_cropped_df <- as.data.frame(CHM_HARV_manual_cropped, xy = TRUE)
Now we can plot this cropped data. We will show the AOI boundary on the same plot for scale.
R
ggplot() +
geom_sf(data = aoi_boundary_HARV, color = "blue", fill = NA) +
geom_raster(data = CHM_HARV_manual_cropped_df,
mapping = aes(x = x, y = y, fill = HARV_chmCrop)) +
scale_fill_gradientn(colors = terrain.colors(10)) +
labs(fill = "Canopy Height") +
coord_sf()
Extract Raster Pixels Values Using Vector Polygons
Often we want to extract values from a raster layer for particular locations - for example, plot locations that we are sampling on the ground. We can extract all pixel values within 20m of our x,y point of interest. These can then be summarized into some value of interest (e.g. mean, maximum, total).
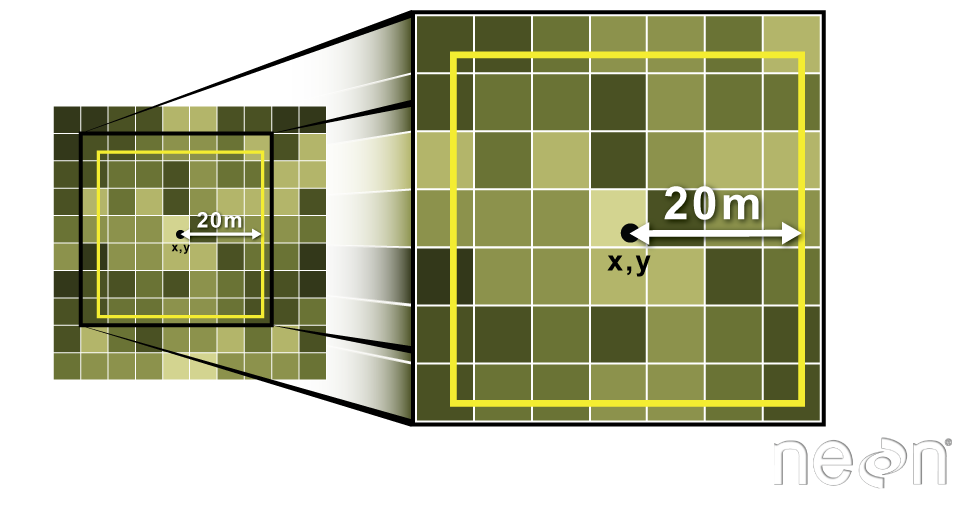 Image Source: National Ecological Observatory Network (NEON)
Image Source: National Ecological Observatory Network (NEON)
To do this in R, we use the extract() function. The
extract() function requires:
- The raster that we wish to extract values from,
- The vector layer containing the polygons that we wish to use as a boundary or boundaries,
- We can also tell it to store the output values in a data frame using
raw = FALSE(this is optional).
We will begin by extracting all canopy height pixel values located
within our aoi_boundary_HARV polygon which surrounds the
tower located at the NEON Harvard Forest field site.
R
tree_height <- extract(x = CHM_HARV, y = aoi_boundary_HARV, raw = FALSE)
str(tree_height)
OUTPUT
'data.frame': 18450 obs. of 2 variables:
$ ID : num 1 1 1 1 1 1 1 1 1 1 ...
$ HARV_chmCrop: num 21.2 23.9 23.8 22.4 23.9 ...When we use the extract() function, R extracts the value
for each pixel located within the boundary of the polygon being used to
perform the extraction - in this case the aoi_boundary_HARV
object (a single polygon). Here, the function extracted values from
18,450 pixels.
We can create a histogram of tree height values within the boundary
to better understand the structure or height distribution of trees at
our site. We will use the column HARV_chmCrop from our data
frame as our x values, as this column represents the tree heights for
each pixel.
R
ggplot() +
geom_histogram(data = tree_height, mapping = aes(x = HARV_chmCrop)) +
labs(title = "Histogram of CHM Height Values (m)",
x = "Tree Height",
y = "Frequency of Pixels")
OUTPUT
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.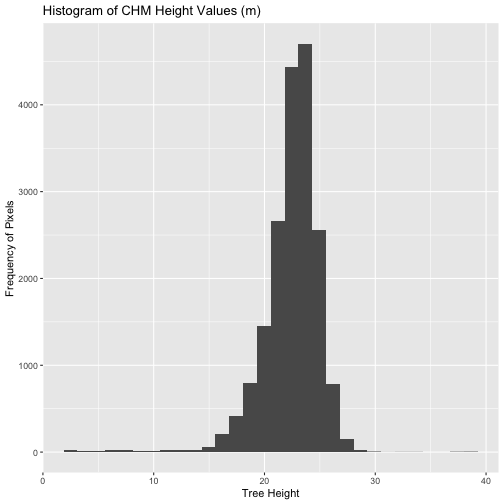
We can also use the summary() function to view
descriptive statistics including min, max, and mean height values. These
values help us better understand vegetation at our field site.
R
summary(tree_height$HARV_chmCrop)
OUTPUT
Min. 1st Qu. Median Mean 3rd Qu. Max.
2.03 21.36 22.81 22.43 23.97 38.17 Summarize Extracted Raster Values
We often want to extract summary values from a raster. We can tell R
the type of summary statistic we are interested in using the
fun = argument. Let’s extract a mean height value for our
AOI.
R
mean_tree_height_AOI <- extract(x = CHM_HARV, y = aoi_boundary_HARV,
fun = mean)
mean_tree_height_AOI
OUTPUT
ID HARV_chmCrop
1 1 22.43018It appears that the mean height value, extracted from our LiDAR data derived canopy height model is 22.43 meters.
Extract Data using x,y Locations
We can also extract pixel values from a raster by defining a buffer
or area surrounding individual point locations using the
st_buffer() function. To do this we define the summary
argument (fun = mean) and the buffer distance
(dist = 20) which represents the radius of a circular
region around each point. By default, the units of the buffer are the
same units as the data’s CRS. All pixels that are touched by the buffer
region are included in the extract.
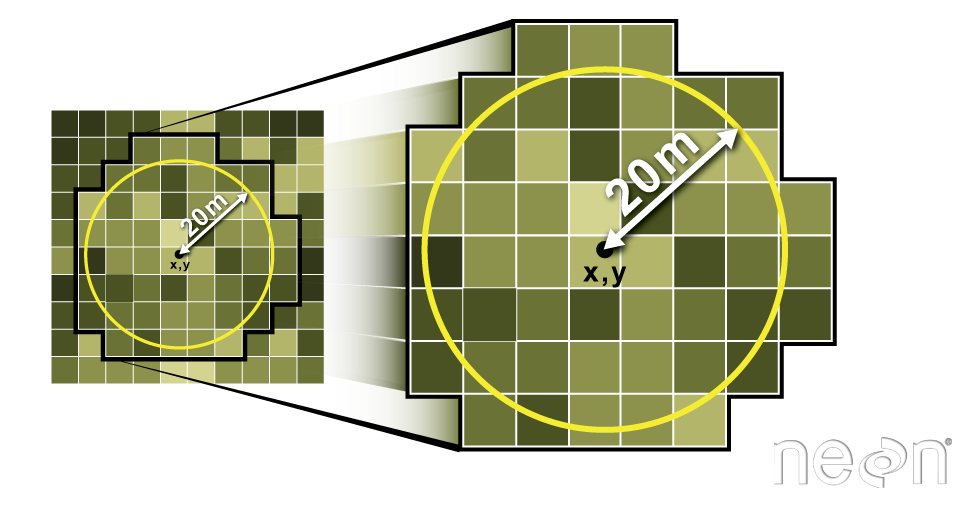 Image Source: National Ecological Observatory Network (NEON)
Image Source: National Ecological Observatory Network (NEON)
Let’s put this into practice by figuring out the mean tree height in
the 20m around the tower location (point_HARV).
R
mean_tree_height_tower <- extract(x = CHM_HARV,
y = st_buffer(point_HARV, dist = 20),
fun = mean)
mean_tree_height_tower
OUTPUT
ID HARV_chmCrop
1 1 22.38806Challenge 2: Extract Raster Height Values For Plot Locations
Use the plot locations object (
plot_locations_sp_HARV) to extract an average tree height for the area within 20m of each vegetation plot location in the study area. Because there are multiple plot locations, there will be multiple averages returned.Create a plot showing the mean tree height of each area.
R
# extract data at each plot location
mean_tree_height_plots_HARV <- extract(x = CHM_HARV,
y = st_buffer(plot_locations_sp_HARV,
dist = 20),
fun = mean)
# view data
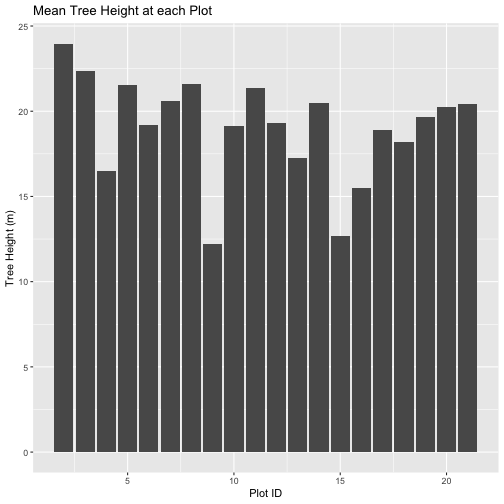
mean_tree_height_plots_HARV
OUTPUT
ID HARV_chmCrop
1 1 NaN
2 2 23.96756
3 3 22.34937
4 4 16.49739
5 5 21.54419
6 6 19.16772
7 7 20.61651
8 8 21.61439
9 9 12.23006
10 10 19.13398
11 11 21.36966
12 12 19.32084
13 13 17.25975
14 14 20.47120
15 15 12.68301
16 16 15.51888
17 17 18.90894
18 18 18.19369
19 19 19.67441
20 20 20.23245
21 21 20.44984R
# plot data
ggplot(data = mean_tree_height_plots_HARV,
mapping = aes(x = ID, y = HARV_chmCrop)) +
geom_col() +
labs(title = "Mean Tree Height at each Plot",
x = "Plot ID",
y = "Tree Height (m)")
WARNING
Warning: Removed 1 row containing missing values or values outside the scale range
(`geom_col()`).